http://www.ebsi.co.kr/ebs/lms/lmsx/retrieveSbjtDtl.ebs?sbjtId=S20180001029
16강 유전 물질(1)
https://hhlife.tistory.com/entry/%ED%83%84%EC%88%98%ED%99%94%EB%AC%BC%EC%9D%98-%EC%A2%85%EB%A5%98-%EB%8B%A8%EB%8B%B9%EB%A5%98-%EC%9D%B4%EB%8B%B9%EB%A5%98%EB%8B%A4%EB%8B%B9%EB%A5%98
================================================================================
Protein
20_kinds_of_amino_acids
several_amino_acids=pick_several_amino_acids(20_kinds_of_amino_acids)
formed_protein=form_protein(several_amino_acids)
================================================================================
DNA
DNA = Nucleotide_1 + nucleotide_2 + nucleotide_3 + ...
================================================================================
Nucleotide
"Ribose" as pentan
 "Phosphoric acid"
"Phosphoric acid"
 "Base"
"Base"
 "Nucleotide"
"Nucleotide"
 ================================================================================
Base=[A,T,G,C]
Nucleotide=[A_based_nucleotide,T_based_nucleotide,G_based_nucleotide,C_based_nucleotide]
================================================================================
Nucleic acid: acidic material in the nuclei
Nucleic_acid=[DNA,RNA]
Unit of nucleic_acid: nucleotide
Nucleotide=form_nucleotide(Phosphoric_acid,ribose,base)
================================================================================
DNA: (-) charge due to phosphoric_acid
Electrophoresis
================================================================================
Base=[A,T,G,C]
Nucleotide=[A_based_nucleotide,T_based_nucleotide,G_based_nucleotide,C_based_nucleotide]
================================================================================
Nucleic acid: acidic material in the nuclei
Nucleic_acid=[DNA,RNA]
Unit of nucleic_acid: nucleotide
Nucleotide=form_nucleotide(Phosphoric_acid,ribose,base)
================================================================================
DNA: (-) charge due to phosphoric_acid
Electrophoresis
 Mixed DNA
Long DNA
Short DNA
================================================================================
Ribose as pentan
Mixed DNA
Long DNA
Short DNA
================================================================================
Ribose as pentan
 Deoxyribose as pentan
Deoxyribose as pentan
 ================================================================================
Base_for_deoxyribose=[A,T,G,C]
Base_for_ribose=[A,U,G,C]
================================================================================
See the structure of ATGCU
Purine_base=[A,G], double ring
================================================================================
Base_for_deoxyribose=[A,T,G,C]
Base_for_ribose=[A,U,G,C]
================================================================================
See the structure of ATGCU
Purine_base=[A,G], double ring
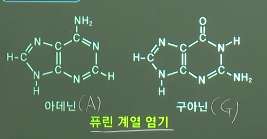 Pyrimidine_base=[C,U,T], single ring
Pyrimidine_base=[C,U,T], single ring
 ================================================================================
Note that base has nitrogen $$$N$$$
So, base is also called nitrogenous base
================================================================================
Elements of DNA: C,H,O,N,P
Pentan: C,H,O
Base: N
Phosphoric_acid: P
================================================================================
RNA=[mRNA,rRNA,tRNA]
mRNA is messanger RNA
rRNA consists of ribosome
tRNA is transfer RNA
mRNA,rRNA,tRNA join the task of creating proten based on DNA
================================================================================
mRNA=copy_specific_part_of_DNA(DNA)
In ribosome (rRNA) which is the place where protein is created:
brought_specific_amino_acid=tRNA_brings_some_amino_acids(amino_acids)
created_protein=create_protein(mRNA,brought_specific_amino_acid)
================================================================================
Places of DNA
- Nucleus
- Chloroplast
- Mitochondria
Places of RNA
- Nucleus
- Chloroplast
- Mitochondria
- Ribosome (rRNA)
- Cytoplasm (mRNA)
================================================================================
Chargaff's rule
Amount of A = Amount of T
Amount of G = Amount of C
$$$\dfrac{T+C}{A+G} = 1$$$
================================================================================
================================================================================
Note that base has nitrogen $$$N$$$
So, base is also called nitrogenous base
================================================================================
Elements of DNA: C,H,O,N,P
Pentan: C,H,O
Base: N
Phosphoric_acid: P
================================================================================
RNA=[mRNA,rRNA,tRNA]
mRNA is messanger RNA
rRNA consists of ribosome
tRNA is transfer RNA
mRNA,rRNA,tRNA join the task of creating proten based on DNA
================================================================================
mRNA=copy_specific_part_of_DNA(DNA)
In ribosome (rRNA) which is the place where protein is created:
brought_specific_amino_acid=tRNA_brings_some_amino_acids(amino_acids)
created_protein=create_protein(mRNA,brought_specific_amino_acid)
================================================================================
Places of DNA
- Nucleus
- Chloroplast
- Mitochondria
Places of RNA
- Nucleus
- Chloroplast
- Mitochondria
- Ribosome (rRNA)
- Cytoplasm (mRNA)
================================================================================
Chargaff's rule
Amount of A = Amount of T
Amount of G = Amount of C
$$$\dfrac{T+C}{A+G} = 1$$$
================================================================================
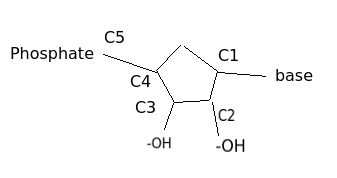
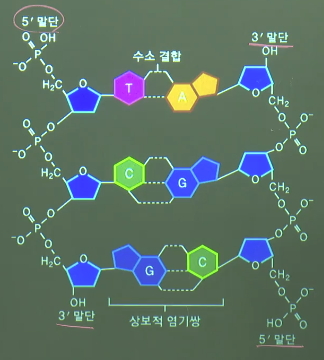
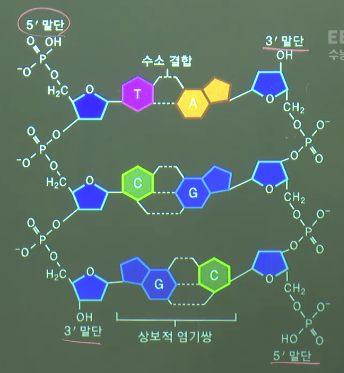 * Phosphate (P) is attached to C5 so it's decribed by 5'
* Phosphate (P) is attached to C5 so it's decribed by 5'
 * You can see base (T) which is coupled to C1
* You can see base (T) which is coupled to C1
 * One nucleotide
* One nucleotide
 * Polynucleotide
* Polynucleotide
 * 2 polynucleotides create DNA
* 2 polynucleotides create DNA
 * Simplified illustration
* Simplified illustration
 ================================================================================
coupled_A_T=double_hydrogen_bond(A,T)
================================================================================
coupled_A_T=double_hydrogen_bond(A,T)
 - double_hydrogen_bond
- single ring (T)
- double ring (A)
coupled_G_C=triple_hydrogen_bond(G,C)
- double_hydrogen_bond
- single ring (T)
- double ring (A)
coupled_G_C=triple_hydrogen_bond(G,C)
 - triple_hydrogen_bond
- single ring (C)
- double ring (G)
Stability of DNA
coupled_A_T $$$\lt$$$ coupled_G_C
================================================================================
temp_AT=Temp_for_separating_DNA_to_single_thread_on_AT_couple
temp_GC=Temp_for_separating_DNA_to_single_thread_on_GC_couple
temp_AT $$$\lt$$$ temp_GC
================================================================================
- triple_hydrogen_bond
- single ring (C)
- double ring (G)
Stability of DNA
coupled_A_T $$$\lt$$$ coupled_G_C
================================================================================
temp_AT=Temp_for_separating_DNA_to_single_thread_on_AT_couple
temp_GC=Temp_for_separating_DNA_to_single_thread_on_GC_couple
temp_AT $$$\lt$$$ temp_GC
================================================================================